 The Denis Furling* team (Paris, France), in collaboration with the Nicolas Sergeant** team (Lille, France), has developed and tested a new approach to gene therapy, a so-called “decoy” approach, for Steinert disease or myotonic dystrophy type 1 (DM1). The strategy is based on the expression of modified proteins that will bind to the pathological expansions of the mutated RNAs in the location of and instead of the endogenous MBNL1 proteins, in order to free the latter and restore their activity in the muscle cells expressing the DM1 mutation. The results have just been published in the Nature Biomedical Engineering*** Journal.
The Denis Furling* team (Paris, France), in collaboration with the Nicolas Sergeant** team (Lille, France), has developed and tested a new approach to gene therapy, a so-called “decoy” approach, for Steinert disease or myotonic dystrophy type 1 (DM1). The strategy is based on the expression of modified proteins that will bind to the pathological expansions of the mutated RNAs in the location of and instead of the endogenous MBNL1 proteins, in order to free the latter and restore their activity in the muscle cells expressing the DM1 mutation. The results have just been published in the Nature Biomedical Engineering*** Journal.
Interview with Denis Furling.
What is the overall context of this project?
DM1 is caused by abnormal expansion of the CTG triplet repeats located in the 3′ untranslated region of the DMPK gene. These mutant DMPK transcripts containing pathological CUG expansions (CUGexp-RNA) sequester, notably, the RNA-binding proteins (RBPs) of the MBNL family, in the form of aggregates or nuclear foci, resulting in the functional loss of the MBNL proteins, leading to defects in RNA maturation and, in fine, gradual deterioration of the muscles. Thus, alternative splicing defects found in the affected muscles of DM1 patients have been associated with clinical symptoms such as myotonia and muscle weakness.
Currently, there is no treatment for this disease, but a number of therapeutic strategies are under development.
What were the objectives of this study?
We have developed a so-called “decoy” approach in order to inhibit CUGexp-RNA toxicity. To this end, an RBP with a strong binding affinity for the CUG expansions was modified in order for it to act as a decoy and prevent the sequestration of the endogenous MBNL proteins by the CUGexp-RNAs.
What methods have you used and what results have you obtained?
We have modified the MBNL1 protein, which has a very strong affinity for the CUG repeats. We have thus generated an MBNL1∆ decoy protein, which has only the RNA binding domains, but highly reduced splicing activity. In vitro competition studies demonstrate that this decoy is able to displace MBNL1 from the CUGexp-RNAs and correct the molecular abnormalities in the isolated muscle cells of DM1 patients.
In order to test its efficacy in a DM1 mouse model expressing CUGexp-RNAs in the muscle, the MBNL1∆ decoy was cloned in an AAV9 vector. Local injection into the muscle of DM1 mice corrects alternative splicing abnormalities, and also myotonia, one of the markers that are characteristic of DM1. This action is long-lasting, since these corrections remain present 1 year after a single injection of AAV-MBNL1∆. The systemic administration of AAV-MBNL1∆ in DM1 mice leads to correction of the splicing defects and an improvement in muscle phenotype.
 What conclusions can be drawn and what opportunities has this study opened up?
What conclusions can be drawn and what opportunities has this study opened up?
Our results underline the efficacy with respect to Steinert’s disease symptoms, of a gene therapy based on the production by bio-engineering of RNA-binding decoy proteins with a strong affinity for pathogenic repeats in the mutated RNA, in order to free the endogenous MBNL1 proteins and restore their regulatory function.
This work is a proof of concept. It will still be necessary to pursue preclinical development, in particular with dose, toxicity, choice-of-sponsor and AAV-vector studies, in order to consider potential clinical applications.
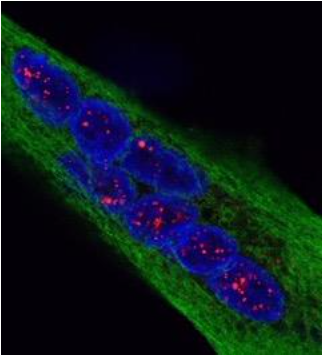
Mutant RNA-DMPK aggregates containing pathological triplet repeats (red)
visualized by FISH/IF in the nuclei (blue) of muscle cells (green)
isolated from patients with Myotonic Dystrophy type 1.
Credits : Denis Furling and Nicolas Sergeant
* Denis Furling, head of the REDs team at the Myology Centre for Research, Inserm/Sorbonne Universiy, Institute of Myology, Paris, France
**Nicolas Sergeant, « Alzheimer & Tauopathies », UMRS1172 Neuroscience & Cognition, Inserm/Lille University, France
